First Person: Building with DNA
By Fenella Saunders
An interview with Yi Lu about DNA nanotechnology
An interview with Yi Lu about DNA nanotechnology

DOI: 10.1511/2014.106.7
Although DNA is best known for its capacity to store genetic information, the double helix has many other remarkable abilities. It can be woven together to build precisely formed physical structures. It can be used to create chemical sensors for environmental monitoring, medical diagnostics, and targeted drug delivery. It can even encrypt messages. Yi Lu is using the material in all of these ways. Lu (below), a professor of chemistry at the University of Illinois and a Sigma Xi Distinguished Lecturer, recently sat down with American Scientist managing editor Fenella Saunders to discuss his innovative blend of genetics, sensing, and cryptography. An excerpt of the conversation follows; a video of the full interview is below, and also available at Sigma Xi’s YouTube channel: www.youtube.com/user/sigmaxisociety

Yi Lu
How did you start using DNA for sensing purposes?
In fact, that was not our initial goal. It was discovered in 1994 that DNA not only can be a regular genetic storage material like we all know, it can also bind metal ions to catalyze reactions. That immediately drew my attention, because as an inorganic chemist I am interested in metal sites in biological systems. So my lab began selecting DNA for a wide variety of different metal ions. Then it dawned on us: Can we utilize this not only for fundamental research, but also for detecting or sensing different metal ions? There are a lot of toxic heavy metals.
What is the link between chemistry and biology in your work?
That’s the core of our research. We can utilize DNA sensors to start asking fundamental questions. Why is iron so good for organisms? How do calcium and magnesium perform their cellular functions? Why is lead so toxic? There is very little information about where lead goes in the body. We don’t know how toxic metals—not only lead but mercury, cadmium, and others—change the function of cells.
How might DNA sensors be used to measure health risks?
One area that we’re interested in is tuning the dynamic range of detection. A good example is lead detection. Detecting lead in water is one threshold, but lead in paint has a completely different threshold. In fact, lead in paint for the wall is one threshold, but lead in paint for a toy is another. Therefore, having a lead sensor is not good enough. We need to vary that dynamic range of detection. We have been able to use different sequences we modified to change the dynamic range.
What about your efforts to improve magnetic resonance imaging?
MRI works beautifully when you have tissue damage or tumors, but when tumors become visible, that’s too late for many people. The new state of the art is detecting biomarkers for cancer before tumors develop. To do that you need MRI contrast differences between the presence and absence of the biomarkers. That’s what we’re developing right now, moving toward noninvasive three-dimensional detection.
You are also designing molecules for drug delivery?
We encapsulate a drug into a liposome, a lipid bilayer, and on the surface of the liposome we put a nucleic acid that is capable of binding to biomarkers for different diseases. The drug will be safer and less toxic, because it is encapsulated, but then when the liposome reaches the target, it can selectively bind to the diseased cell and not the normal healthy cells, and dump its drug in there.
You also use DNA as a building material. How does that work?
The key word of DNA is programmable. Not only can it form a double helix, it can form a triple helix, or even three-dimensional shapes. An exciting field is DNA origami: We can program the DNA sequences to fold into whatever shape you want. Multiple strands of DNA can form cubes, cylinders, faces, maps, flags, curved surfaces, corners, and different angles. It’s really exciting. DNA gives you shape control that is difficult to do with other methodologies.
Why are you so excited about being able to un-build structures as well?
There has been very limited research in reversible assembly—how to take parts out and put them back in, in a reversible way. But if you look at biological systems, DNA and proteins do this all the time with transcription and translation. One component goes in, it goes and does a job, then when it finishes the work it goes back and a second component comes in. Inspired by the biology, we can learn how to do that. In fact we use a lot of the fundamental parts of the biological system. We use [the enzyme cofactor] biotin and its derivative desthiobiotin; they have slightly different affinities for [the protein] streptavidin, which allows us to do nanoscale assembly in a reversible way.
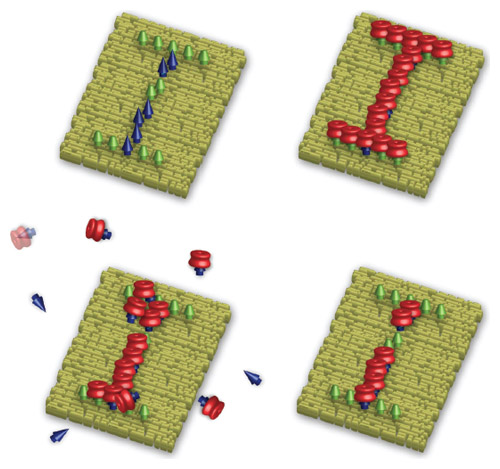
Images courtesy of Yi Lu.
What is the connection between reversible assembly and data privacy?
One application of reversible assembly is encrypted messages. In the spy game, you send somebody a piece of paper with no messages, or random messages, then they add invisible ink to it and the real message appears. This is what we were able to demonstrate, but it’s a nanoscale spy game now. Embedded in a nanometer-size square of DNA origami is the message N-A-N-O, using Morse code. We cover that with another type of material so when you receive it you will have no message. When you add an enzyme to it, it can replace the weaker binders and act as the invisible ink. The ones that remain, the strong binders, are coded for N-A-N-O. But you need a microscope to see that message. [See the video below for narrated images of this research.]
Where do you see this technology going in the future?
One major focus for us is to make the DNA metal sensor into a cellular detector so we can really start telling people why lead is so toxic, where does it go, what are its effects. Calcium’s the same thing: In the different stages of development, why does calcium work in different places? The second thing we’re interested in is more fundamental. Why is this particular DNA sequence so selective for lead? Once we know more about the features responsible for binding these metal ions, we can design smaller and more effective sensors. Another area of research is to expand the scope of the detection. Every year there’s a new type of flu. Can we use this methodology to quickly select different DNA to target different ones? So far most of our work has been using regular nucleic acid. Why not combine an amino acid with a nucleic acid? Then you can have the functionality of both, and the ability to bind to a target better, tighter, and more selectively.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.