When PPLO Became Mycoplasma
By Harold J. Morowitz
The smallest cell has had a long career in the spotlight
The smallest cell has had a long career in the spotlight

DOI: 10.1511/2011.89.102
Now that Craig Venter, Hamilton Smith and Clyde Hutchison, visionaries in the attempt to synthesize a living cell, have created three species of mycoplasma into landmarks on the road to their ambitious goal, mycoplasmas have become a favorite taxon in the popular press and the world of futuristic biology. When I first encountered these organisms they were known as pleuropneumonia-like organisms—PPLOs—and few dreamed of the glory that lay ahead for this enigmatic group. How did these obscure microbes became important, indeed central, in understanding what it takes to be a living organism? The story will be told from an idiosyncratic and personal point of view—the view from my bench over five decades—and will show how mycoplasma instructed—and deceived—us about the nature of life.
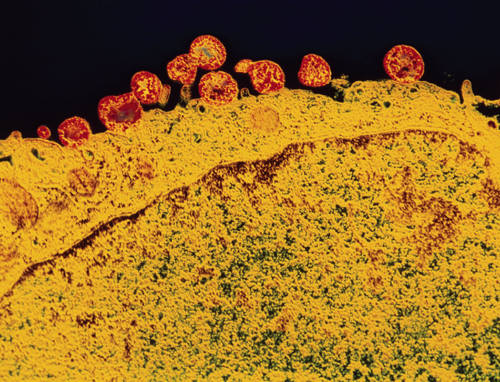
CNRI/Photo Researchers
As an assistant professor of biophysics at Yale University in the late 1950s, I decided to focus on the origin of life, a subject that had intrigued me since I had read Erwin Schroedinger’s profound little book, What is Life? Agreeing with da Vinci that simplicity is the ultimate sophistication, I decided to look for the smallest and simplest free-living organism, thinking it would be the closest to the earliest organisms. Serendipity struck and Bob Cleverdon, professor of microbiology at the University of Connecticut, appeared in my office and announced that he was looking for a laboratory at Yale to spend a one-year sabbatical. Bob was a broadly trained and experienced bacteriologist, and the search for the smallest and simplest free-living microbe struck his imagination.
Shortly thereafter our collaboration began. Bob wrote letters to a large group of bacteriologists he knew, and I took to Bergey’s Manual of Determinative Bacteriology and the American Type Culture Collection looking for species with words like parvulus in the species name, hinting at tiny cells. Soon cultures were arriving by mail, and the incubator became filled with flasks and petri dishes. The search was complicated by the fact that the smallest bacteria were near the limit of visibility using optical microscopy, and we were not at all sure about what criteria to use as the measure of simplicity. While we were pondering this, the DNA genome became accepted, and the DNA content—total weight and ratio of nucleotides—of several bacterial genomes were appearing in the literature. It seemed obvious to us that simplicity was the same as the smallest amount of DNA in the bacterial chromosome. At that time the genome and the single bacterial chromosome were considered to be the same. Our task changed to seeking the organism with the smallest amount of DNA in the postdivisional cell.
At this juncture serendipity struck again when Cleverdon became aware of the work on pleuropneumonia-like organisms being done by Mark Tourtellotte, a graduate student in the Department of Animal Diseases at the University of Connecticut College of Agriculture. A conversation between Bob and Mark informed us that PPLOs could pass through the fine filters that captured normal bacteria and hence were probably very small, although the fact that they lacked a rigid cell wall might indicate that they were sufficiently deformable to ooze through the filter. In any case it was clear that PPLOs had to be added to our collection of candidate organisms and that a new array of laboratory techniques would have to be learned.
Eventually Bob’s sabbatical was over and he returned to the University of Connecticut, passing the baton to Mark Tourtellotte, who was traveling in the opposite direction from Connecticut to Yale for a post-doctoral fellowship. The smallest-genome project now had two nodes operating cooperatively.
Serendipity struck again when Melvin Calvin came to visit our department and was brought to our laboratory by the department chair, Ernest Pollard. Professor Calvin had won the Nobel Prize for his work unraveling the famed Calvin cycle at the heart of photosynthesis. When we met him, he was the chief consultant to the newly formed biology division of the National Aeronautics and Space Administration (NASA). Upon hearing of our search for cellular simplicity, he noted, “that’s just the kind of thing that NASA should be interested in.” This encounter resulted in grant support for our work for the next 15 years or so. Shortly after our entry in this new field, a paper appeared by Morowitz and Cleverdon entitled “An extreme example of the coding problem, Avian PPLO 5969.“ Using the data and techniques available at the time, we calculated that there were only 70 protein-coding genes in our little cell. And that was assuming the DNA was double-stranded, which we suspected but did not know at the time. (By modern estimates, the number of protein-coding genes in PPLO 5969—Mycoplasma gallisepticum—is not 70, but about 865. As we’ll see, it wouldn’t be the last time mycoplasmas fooled us.)
To understand the problems we encountered in trying to determine the genome size of PPLO, it is necessary to look into the background of this group of microbes. In the 1890s, two French investigators, Edmond Nocard and Emile Roux were studying pleuropneumonia in cattle. This was a disease of economic importance. Both Nocard and Roux had been trained by Louis Pasteur, taking us back to the beginning days of bacteriology. In 1898 they published the first report of mycoplasmas in a paper, “Le Microbe de la péripneumonie.“ To indicate what was special about these organisms we quote from Emmy Klieneberger-Nobel, the grande dame of the small community of PPLO researchers at the time we began our studies.
All this started in 1898 with a remarkable discovery by French bacteriologists, Nocard and Roux, namely that pleuropneumonia, a highly infectious disease of cattle, was caused by an organism that could pass filters that retained bacteria. It was at first not possible to cultivate this organism on customary bacteriological medium: but the authors soon produced a richer medium on which this organism, seen in their microscopes as little dots darting around in Brownian motion grew in the form of tiny delicate colonies. These were characterized by a dark center and lighter peripheral zone.
This brief description of very small size and growth outside of a host recalls the beginning of PPLO fame as the smallest living organism. Filtration placed them with the viruses, and independent growth classed them with the bacteria.
Twenty-five years passed before any other organisms of this type were discovered. Then in 1923, J. Bridre and A. Donatien reported on an organism similar to the causative agent of pleuropneumonia in cattle appearing as the causative agent of agalactia (faulty secretion of milk) in sheep. By the early 1930s a number of similar organisms were found, largely linked to animal diseases, and they were conveniently referred to as pleuropneumonia-like organisms following the first member of the group so discovered. Work on the group picked up in many countries, largely pursued by veterinary pathologists. In 1937, the first human disease, a pneumonia, was shown to be caused by a PPLO. And so PPLO moved into mainstream bacteriology, led by investigators such as Klieneberger-Nobel at the Lister Institute of Preventive Medicine in London. At that time a complication arose, which in retrospect was due to PPLOs being organisms that lacked a cell wall. Normal bacteria, which possessed a cell wall, could enter a phase where they lost their cell walls but were stable if the osmotic pressure of the medium was high enough. Cells in this stage were designated L forms and were generally capable of reverting to the walled condition. Because PPLOs lacked cell walls, there was 30 years of confusion as to the relation of PPLO to bacterial L forms.
When we undertook to study PPLOs it was not clear that they were normal cells surrounded by the usual cell membrane. They lacked cell walls, were so small they could barely be visualized with optical microscopy, and they had a tendency to clump in many strange ways. Cellularity and mode of replication was uncertain. The growth media used were complex, requiring a mishmosh of components such as blood serum to induce these balky creatures to grow. Drying and embedding specimens for examination by electron microscopy caused so much distortion that one could hardly rule between reality and artifact. All of this added to the problem of determining the size of the genome. On the plus side, a number of graduate students at the University of Connecticut and Yale joined the game, so there was a lot of help in tackling the problems.
In January 1959 a conference on Biology of the Pleuropneumonialike Organisms was held by the New York Academy of Sciences. There were about 200 attendees with speakers from nine countries. The meeting was chaired by D. W. Edward of the Wellcome Research Laboratory in England. The speakers included most of the pioneers, largely from veterinary and agricultural institutions, who were struggling to understand organisms that did not fit the paradigm of bacteria or viruses. Another feature appeared at this juncture. Tissue-culture technology had come into widespread use and the media were often sterilized by filtration. Pleuropneumonia-like organisms were starting to show up frequently as tissue-culture contaminants. When we began culturing PPLOs at Yale, I recall frequent phone calls from laboratories around the university that were growing tissue cultures. Some were requests for information on how to avoid PPLO contamination and some were accusatory, as if our sloppy ways were spreading these organisms around New Haven.
We plowed ahead trying to measure the amount of DNA and the number of colony-forming units in cultures under varying conditions. We continued using the strain of avian pathogen known as A5969. Jack Maniloff began detailed studies of this strain using electron microscopy. Word got around that PPLOs were among the smallest of genomes and this proved of interest to a number of investigators. I recall meeting Leo Szilard, the famous nuclear physicist turned molecular biologist, at a meeting of the American Physical Society in Washington. On being introduced and hearing my name he asked, “What is the size of the PPLO genome?” Although we had values, the uncertainties about morphology, cellularity and mode of replication left the meaning of these values uncertain.
Our discussions suggested a method of confirming cellularity by means of predictions about closed vesicles made of membranes of known high impedance. This allowed us to use a technique discovered 100 years earlier. James Clerk Maxwell, in the early days of electromagnetic theory, worked on the problem of the impedance of a suspension of conducting spheres surrounded by nonconducting shells and embedded in a conducting medium. In the 1920s this theory was tested for biological materials by placing a suspension of red blood cells between two platinum plates and measuring the impedance as a function of frequency of the applied field. From the values obtained, one is able to calculate the inductance of the nonconducting shell. With these results and a knowledge of the dielectric constant of the shell, arrived at by assuming it was made of lipids, a value was obtained for the thickness of red blood cell membranes. This was the first experimental determination of this important biological parameter.
We reasoned that if PPLOs were vesicular in nature, we should be able to demonstrate it to be so. A center for work on dielectric dispersion was the Moore School of Engineering at the University of Pennsylvania under the direction of Herman Schwan. I knew Schwan from various meetings of biophysicists and made contact to carry out the necessary experiments. And so with a Styrofoam cooler packed with ice and a number of pellets of PPLO A5969, I made my way from New Haven to Philadelphia for several days of intensive experiments with my engineering mentor. The result was a paper, “Electrical properties of the membranes of pleuropneumonia-like organism A5969,” and a conviction that the cells were indeed vesicles surrounded by the usual type of cell membrane.
There was some confusion in the 1950s as to the name of the taxon that contained PPLOs. In 1958 Eyvind A. Freundt of the Statens Seruminstitut in Copenhagen, one of the world authorities on this group of organisms, authored a monograph, The Mycoplasmataceae (The Pleuropneumonia Group Of Organisms). Freundt’s paper a year later in the 1959 New York Academy Symposium was entitled “Morphology and classification of the PPLO.” I stress the confusion in the early days as to the morphology, physiology, phylogeny and nomenclature of the organisms that looked like the best bet in our search for smallness and simplicity. What was most difficult is that we had no idea of how this group of organisms related to other microorganisms, by phylogeny or biochemistry. Another enigmatic feature of the PPLO was its requirement for cholesterol, which was generally considered to be exclusively a constituent of animals and not seen in bacteria.
In 1962 Bob Cleverdon arranged an extensive workshop on PPLOs at the University of Connecticut. It brought us in contact with Shmuel Razin, who was building a center for PPLO research at the Hebrew University Medical School in Jerusalem. Razin, who came to Connecticut for a sabbatical the following year, was for the next 40 years a central figure in PPLO studies, presiding over his own center and traveling and collaborating with researchers around the world. There is hardly an area in mycoplasmology in which his influence is not felt. His insights contributed to our growing confidence in working with these strange microbes. By 1962 Mark Tourtellotte and I felt confident enough to write an article for Scientific American entitled “The smallest living cells.”
The period from 1962 until 1966 was characterized by a worldwide effort to promote the PPLO from veterinary pathogens to microbes that were understood in terms of biochemistry, morphology and growth characteristics. This was also a period when molecular biology and the genetic code were coming of age. In 1966, The New York Academy of Sciences decided to revisit this group of organisms in a conference entitled “Biology of the Mycoplasma.” The linguistic transition from Pleuropneumonia-like Organisms to Mycoplasma was now complete. Mycoplasma were cellular microorganisms lacking a cell wall but bounded by a cell membrane. The membranes contained cholesterol, which was not synthesized by the mycoplasma but was obtained from the host or from the growth medium. The conference had over a thousand attendees.
At this time our laboratory focused on the size and replication pattern of the mycoplasma genome. The problem was elegantly solved in the dissertation work of Hans Bode, now professor at the University of California, Irvine. He spent the rest of his career after getting his Ph.D. as one of the world leaders in hydra morphogenesis. (Given the current focus on the mycoplasma genome, Bode’s important early work has been improperly overlooked.) In the 1961-to-1964 period, extensive work was being done in several laboratories on a number of bacteria using radioautography and a protein-film electron-microscopic technique developed by Alfred Kleinschmidt and Rudolf Zahn. Using cultures of Mycoplasma hominis sp. H39, cells were lysed on an air-water interface while spreading in a protein film. Samples were then transferred to Formvar grids, dried and shadowed with platinum and palladium. They were then examined in the electron microscope and lengths determined. From the lengths and calibration with phage DNA, the genome size was shown to be 510 million daltons. Work on a number of other strains gave genome values in the range of 450 to 1,000 million daltons. Bode’s work was published in 1967 in “Size and structure of the Mycoplasma hominis H39 chromosome.” Mycoplasma had the smallest genome of any microorganism known at that time.
In the late 1960s Carl Woese was revolutionizing taxonomy based largely on the sequences of ribosomal RNA molecules. When he and Jack Maniloff looked at mycoplasma in this context, it became apparent that the organisms were descended from bacteria that had lost their cell walls and taken up a parasitic existence. The lush environments to which they became adapted—such as the humid greenhouse environs of the human lung—allowed them to shed swaths of their genome and metabolic capacity, living instead as obligate parasites dependent on biomolecules synthesized by their hosts.
From serendipity to implacable reductionism: Our original reason for studying PPLOs, to learn what there was to be learned about the original cells at the origin of life, had been completely misdirected. Rather than being an early taxon, it was a late one—very late. Among the newest of the microbes. It had lost size and genome due to its life in hosts of the Cambrian or later. While I try to think of a moral for this story, I’ll say in an aside: Sorry, Dr. Calvin. Mea culpa. In another bit of serendipity, Carl Woese, a coauthor of one of our earliest PPLO papers, developed the technique and scientific framework that solved some of our early confusion about who the mycoplasma really were. This nucleotide-sequence work was long after I had left the mycoplasma genome and turned attention to the mycoplasma membrane, another source of fascination offered up by this organism.
In 1980 Mycoplasma genitalium was isolated from a case of non-gonococcal urethritis. By then I had left this field of research. Sometime thereafter, Clyde Hutchison of the University of North Carolina began stalking the smallest genome, including sequencing its DNA. In 1995 a group at the Institute of Genomic Research headed by Claire M. Fraser finished the complete sequence of the Mycoplasma genitalium genome. Among the coauthors of the published report were Clyde Hutchison, Hamilton Smith, and Craig Venter. Mycoplasma has moved in my mind from one of the oldest to one of the most recent microbes. Indeed, in the hands of Venter and his crew it is moving into the future. If they are fully successful and start seeking a name for their entities, perhaps they might call them PPLOs—patent-pending laboratory organisms.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.