Salivary Diagnostics
By David T.W. Wong
Amazing as it might seem, doctors can detect and monitor diseases using molecules found in a sample of spit
Amazing as it might seem, doctors can detect and monitor diseases using molecules found in a sample of spit

DOI: 10.1511/2008.69.37
Saliva carries different meanings around the world. In most of the United States, for example, the act of spitting is taken as an insult, whereas in some other cultures it can be considered a blessing.

Paul DeVay
And although most Americans do not have a problem with the occasional wet kiss on the lips, they react with revulsion when they see objects covered with saliva. This seeming double standard arises because people perceive a difference between the saliva in their mouths and the saliva outside their bodies, according to Gordon Allport, a Harvard psychologist who penned an influential 1960 publication on the subject.
People won't drink their own saliva, explained Allport, who hypothesized that this fluid becomes nonself and alien to the mind the moment it exits the mouth. Maybe this aversion explains why the biomedical community has been slow to recognize that saliva doesn't just help one to chew and swallow—it also contains information about the physiological states of the body.
Like blood, saliva contains many protein and RNA molecules, both of which are encoded by genes. Scientists can identify several abnormal conditions if they know which genes are active and at what levels—information that can in many instances be gleaned from a sample of a person's blood. Saliva, however, is far easier and cheaper to collect and doesn't expose health-care workers to blood-borne diseases. Oral fluids are also simpler to handle because they don't clot, lessening the manipulations required. Furthermore, it's possible that diagnoses that use saliva could be made outside of a doctor's office, which is attractive for people who can't afford to see a physician or for people living in places where there are none.
Several tests that use saliva are already on the market. Three years ago, the U.S. Food and Drug Administration approved a product called OraQuick for detecting HIV-1 or HIV-2 infection. The assay, which is sensitive to anti-HIV antibodies in oral fluid or blood, indicates the result with one or two colored lines, similar to a home pregnancy test. The FDA authorized the use of OraQuick in clinical settings, but future versions may be sold over the counter. Commercially available kits can gauge the levels of a handful of hormones, including estrogen, testosterone and cortisol, from a sample of saliva. And other, yet unapproved, methods can screen for hepatitis viruses.
In addition to these simple measures, saliva has the potential to diagnose diseases with more complex origins, including cancer and diabetes. In recent years, my colleagues at the University of California, Los Angeles, and I have studied the RNA and protein molecules in saliva as indicators of disease. We've found that we can diagnose early-stage oral cancer and Sjögren's Syndrome, a systemic autoimmune disease marked by dryness of the mouth and eyes. Salivary diagnostics for serious illnesses affecting other parts of the body may be just around the corner. But to understand how biomedical researchers can perform such feats, one must first have a clear understanding of the makeup of this remarkable bodily fluid.
Saliva is mostly water, but it also contains protein molecules that lubricate our wagging tongues, inhibit the growth of bacteria, prevent excessive swings in pH and begin the process of digestion. Unfortunately, the importance of saliva is often appreciated only when it's gone, as commonly happens in patients who get radiation treatments or have oral cancer. These individuals routinely face speech problems, and simply chewing and swallowing everyday foods becomes difficult. Without saliva, the mouth is vulnerable to bad breath, yeast infections, cavities and gum disease.
Stephanie Freese
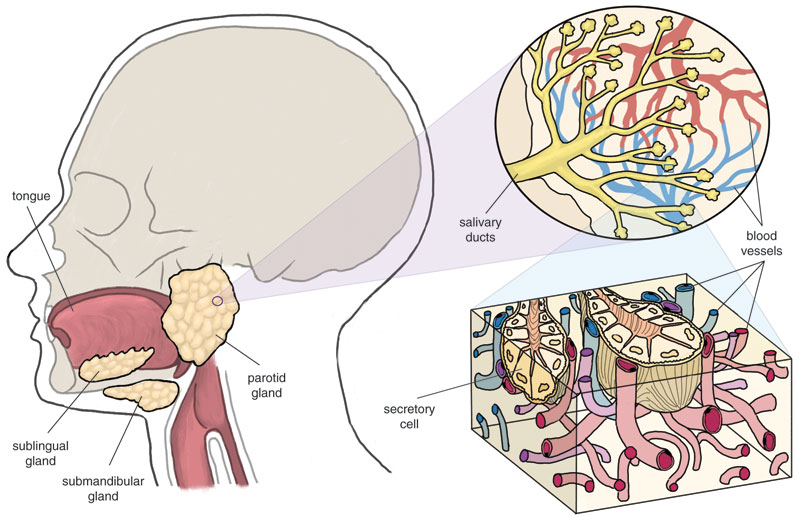
Saliva comes primarily from three distinct places—the parotid, the submandibular and the sublingual salivary glands, where specialized cells take up water, salts and macromolecules from the blood, mix them with a cocktail of saliva-specific proteins and secrete the resultant concoction. Some substances may also reach saliva by passing from blood through the spaces between cells. Hence, most compounds found in blood are also in saliva, leading to the aphorism among dentists that saliva is the "mirror of the body." It's a mirror that reflects the levels of natural and artificial substances, including drugs taken for therapeutic or recreational purposes. Saliva can also indicate emotional and hormonal status, the health of the immune system, neurological conditions, nutritional deficits and metabolic states.
Currently, most molecular diagnoses are based on blood samples, and for good reason: Serum, the cell-free, liquid component of whole blood, typically contains high concentrations of the molecules of interest. But newer, more sensitive tests enable the detection of very small amounts of material. This advance in technology has reduced the importance of having a high level of the target compound.
Photograph courtesy of the National Institute of Dental and Craniofacial Research.
One of the most important reasons for developing saliva-based diagnostic tests is a matter of simple economics. In situations where saliva and blood can both serve, it might make sense, from the patient's perspective, to use blood—after all, the quantities of most biomarkers are higher in blood than in saliva. The momentary discomfort seems a small price to pay for ruling out some rare, undetected disease. But for insurance companies, it may be that only saliva-based tests are inexpensive enough to use in a large population. If the condition is sufficiently rare, treating the late stages in the few people that develop it costs less than checking everybody for early signs. This seemingly cold-hearted calculation is, of course, something insurers are very sensitive to.
Were cost not a consideration, it would always be smart to screen people for both common and uncommon diseases. After all, even with a physician's most astute observations and modern laboratory tools, rock-solid diagnoses early on are not the norm, and many diseases remain hidden until they become quite advanced. For this reason, medical researchers are keen to identify biomarkers of disease: molecules, usually DNA, RNA or protein, that act as proxies for particular physiological states. Physicians could then use these indicators to discover sicknesses, perhaps even before the onset of symptoms. The best biomarkers are both specific and reliable, meaning that they are uniquely indicative of a particular disease and that all the people with that illness have the markers.
Barbara Aulicino and Stephanie Freese
Although the benefits of diagnostic molecules are obvious, relatively few have been approved for use in the clinic. The modest number is not from inattention—scientists in academia, government and industry have devoted significant resources to finding the molecules that signal disease. In particular, the National Institute of Dental and Craniofacial Research has invested heavily in research to ascertain which conditions might reveal themselves through components found in saliva. However, biological systems have proven more complex at the molecular level than I and other investigators thought even a few years ago. Specific physiological states seldom reveal themselves with a change in one protein or RNA. Instead, the molecular hallmark of a disease may consist of altered levels of RNA from many genes—more than a hundred in some of our preliminary studies. In such cases, no single marker can form the basis of a diagnosis, but many, taken together, can yield a more detailed picture of a person's physiology.
Looking for patterns among the variations of thousands of potential biomarkers is a daunting task and one that requires much basic study before approaching the point where disease signals can be routinely detected. For that reason, my colleagues and I at UCLA began a few years ago to catalog the entire collection of proteins and RNAs found in the cell-free portion of saliva. Fortunately, techniques refined in the past 10 years have made it possible for scientists to examine many RNA or protein molecules at once, making our ambitious quest feasible.
In 2003, with funding from the National Institute of Dental and Craniofacial Research, our team began studying the proteins in saliva from healthy individuals. We first split each of the samples and then studied the different parts using different techniques. In each case, we separated the proteins into ordered groups, or fractions, according to some physical attribute of the molecule, such as size or charge. Then we analyzed the fractions independently. This "divide and conquer" strategy reduced the complexity of the protein mixture and provided a measure of redundancy (and thus validation) for our results.
The goal of this study was twofold: first to identify the individual proteins present in the saliva, and second, to get an idea of how abundant they are. While some members of my laboratory are continuing to pursue this whole-proteome initiative, other team members are creating a catalog of glycoproteins, or proteins that have attached sugar molecules. Such sugars decorate many of the proteins most important for the proper functioning of saliva, including mucin, which protects and lubricates the lining of the mouth and throat, and amylase, which breaks starch molecules into glucose. In this case, an analytical technique called liquid chromatography-tandem mass spectrometry helps us to identify the specific types of sugars and their sites of attachment on the protein.
As of November 2007, we have cataloged more than 1,000 proteins in whole saliva. To help disseminate these results, we developed an online database that contains our accumulated knowledge of these proteins. This Web site is freely available to the public, provides a repository for data and allows us to compare our results with information from other protein databases. We've learned that many of the proteins we find in saliva have already been discovered elsewhere in the body. The investigators who carried out these earlier studies often assigned them names and described their functions.
As we have accumulated information about the proteins in saliva, an ongoing project of ours has been to examine carefully the differences between the human salivary proteome and the collection of proteins found in human plasma. The molecular constituents of these two fluids are not identical. Indeed, our preliminary efforts show that proteins found in saliva tend to be more hydrophilic (which is to say that they are attracted to water molecules), whereas the proteins in plasma were more often hydrophobic. When we looked at known proteins from each group, we noted that extracellular proteins—those that are normally excreted into the spaces between cells—are more abundant in saliva than they are in plasma. By contrast, proteins that are known to reside in or near the lipid membranes of cells are found more often in plasma than in saliva. These observations are consistent with the fact that saliva is a filtrate of blood, meaning that the contents of blood—water, salts and macromolecules—pass through capillary walls into the salivary glands. And they can sometimes provide hints about a molecule's function, because what a protein does is often tied to where in the cell it is found.
Members of my laboratory discovered a few years ago that, much to everyone's astonishment, many RNA molecules, or transcripts, as they're often called, are present in the cell-free portion of human saliva. These transcripts include messenger RNA or "mRNA," the kind of RNA that cells use to convey the instructions carried in DNA for the manufacture of proteins. This finding was surprising because RNA tends to be quite fragile outside the confines of the cell. Indeed, we don't know for certain where this RNA comes from. Cells do not ordinarily secrete this type of molecule, so we suspect that it comes from leaky or broken cells. The makeup of the RNA population doesn't suggest that it originates from a single type of tissue, which leads us to conclude, for now, that such leaks or breaks may not be unusual considering the quantity of cells in the human body—a number that most experts estimate to be in the trillions. In addition to proteins, the collection of RNA molecules constitutes a second saliva-based diagnostic alphabet for disease diagnosis.
Zephyr/Photo Researchers, Inc.
In a 2004 publication, we noted that saliva contains approximately 3,000 mRNAs, of which 180 were common between samples from 10 healthy people. This slight degree of overlap (17 percent) might be considered low for cells, all of which require a common set of "housekeeping genes" to provide their basic building blocks and to enable routine metabolism. However, because we studied the cell-free portion of saliva, we weren't particularly surprised by the results. Nonetheless, expecting the variation and explaining it are quite different propositions, and we cannot say why the large majority of RNAs from our study subjects were different. We suspect that this aspect of salivary function, similar to so many other physiological differences between one person and the next, will prove to be a product of both genes and environment.
Having established an RNA baseline for normal saliva, we began searching for RNA biomarkers that could help diagnose disease. Our first target was oral cancer, the most common form of cancer in developing countries. In India, for example, oral cancer accounts for 40 percent of all malignancies. The incidence is lower in the United States, where oral cancer makes up less than 3 percent of the total. Nevertheless, more than 28,000 new cases were diagnosed in this country in 2004. Tobacco use, whether it is smoked, chewed or held in the mouth, represents the primary risk factor for cancers of the mouth and pharynx. Drinking alcohol also raises the risk in a dose-dependent fashion, but to a much lesser extent than does tobacco. Additionally, some studies implicate certain human papilloma viruses in the genesis of these tumors. However, more than a quarter of oral cancer victims neither smoke nor drink and have no other lifestyle-related risk factors.
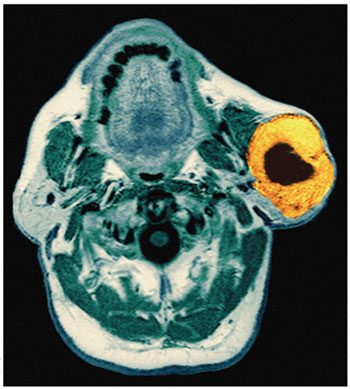
Oral cancer typically begins as a lump within the mouth, but finding it is not always straightforward. In the early stages, such growths are hidden more often than not, making visual inspection an unreliable means of diagnosis. Many of these abnormal tissues, visible or invisible, will never become cancerous. Furthermore, it can be difficult, even for experts, to distinguish between benign tumors and early-stage malignant ones. Of course, the most reliable means of diagnosis, a surgical biopsy, is also the most invasive and is unsuited for use as a screening technique. As a result, several other means of identifying this type of cancer have emerged in recent years. Some, such as illumination with certain wavelengths of light (autofluorescence) or application of the dye toluidine blue, detect higher concentrations of RNA and DNA in clusters of cancerous cells, which are often crowded together compared with normal cells. Suspicious areas can then be examined further with a biopsy. (Autofluorescence has already entered clinical practice in the United States; toluidine blue is in phase-III clinical trials and has been approved for use in Europe.) Another new technique uses a combination of vinegar, dye and special lights to heighten the ability to see lesions in the mouth. It's also possible for scientists to search under the microscope for cancer cells in tissue from a scrape of the cheek.
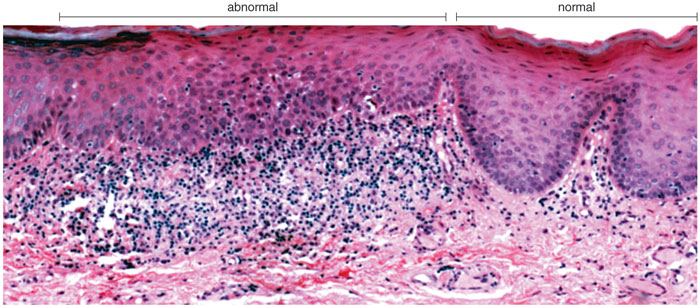
Photomicrograph courtesy of Nadarajah Vigneswaran, University of Texas Dental Branch at Houston.
These techniques are generally effective at identifying malignancies. However, they have some disadvantages. Principally, they are all limited by the need to examine the external surfaces of the mouth, which must miss some proportion of early-stage cancers. In addition, and perhaps more important from a public-health standpoint, none of these technologies promises to become inexpensive enough to screen people in the general population, because they all require expert medical personnel to conduct the test and judge the outcome.
I hope that a saliva-based test will one day provide an improvement over any of these methods. The trick, of course, is to identify the telltale signs of oral cancer in saliva. In a search for RNA biomarkers, my colleagues and I compared the transcripts found in the saliva of people with early-stage oral cancer with those of people without cancer. This work involved a technique called microarray analysis, a type of test that allows the simultaneous measurement of thousands of RNAs.
In a microarray experiment, scientists attach a kind of molecular beacon to all the RNAs in a particular sample and then see what DNA sequences they attach to. (An RNA molecule and its mirror image, or complement, DNA bind tightly to each other.) The microarrays are palm-sized quartz wafers on which millions of DNA molecules are laid out in a grid, or array. There are about 22,000 different sequences among these short pieces of DNA, each corresponding to a different RNA transcript. The same sequence is present at many locations on the grid. This redundancy helps eliminate false signals and provides an internal check. After the tagged RNAs find their DNA complements, a laser "reads" each position on the array and indicates whether those RNAs are present in the sample and in what quantities.
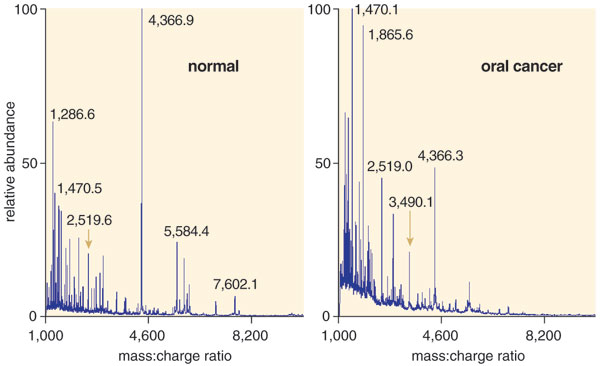
Using this approach, we examined the salivary RNA from 32 patients with early stage oral cancer and 32 controls and found that four of the 180 common transcripts showed consistently different levels in cancer patients. Among them, 91 percent had the distinctive changes in these four RNAs, confirming that the markers are reliable indicators of cancer. When we searched a larger group of people, we found that very few people had these four biomarkers. But among those that did, more than 9 out of 10 had oral cancer. The next step will be to conduct a similar study on a wider scale. To date, we've confirmed the findings in more than 300 oral-cancer patients, and the National Cancer Institute's Early Disease Research Network has validated our results.
In addition to our investigations of oral cancer, my research team and our collaborators have worked to identify salivary biomarkers for several other diseases. Most recently, we published a paper on a group of 26 RNA biomarkers that characterize Sjögren's syndrome, which primarily affects the salivary glands and tear ducts but can also cause symptoms in many other tissues. Although it is not well known, Sjögren's syndrome is a common condition, affecting between 1,000,000 and 4,000,000 million people, predominantly women, in the United States. The wide range of this estimated prevalence reflects the difficulty of identifying this condition. As with many autoimmune diseases, symptomatic episodes come and go, varying in duration and severity. The Sjögren's Syndrome Foundation reports that the average patient experiences a lag of six years between the onset of symptoms and diagnosis.
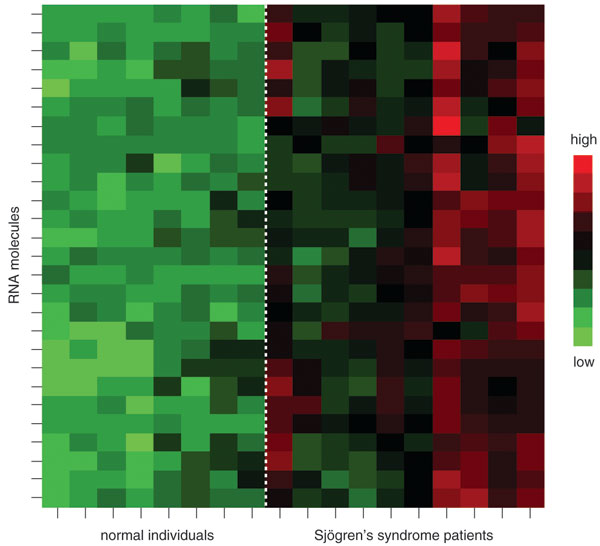
My colleagues and I have also focused considerable effort on finding RNA signatures for major diseases that affect parts of the body other than the mouth. Although we have not published these results, our preliminary studies identified a group of RNA biomarkers that appear to characterize breast cancer. We've also found evidence that salivary biomarkers may support the diagnosis of type-II diabetes and pancreatic cancer. However, it is important to note that these studies are ongoing.
Another forward-looking project that we are excited about is a collaboration with Chih-Ming Ho in the School of Engineering at UCLA. Our colleagues there have developed so-called "micro- and nano-electrical-mechanical-system" biosensors—tiny, self-contained machines that automate the process of detecting extremely small quantities of target molecules. Within a few years, we hope to have prototypes for a handheld device that would enable an investigator or clinician to detect proteins or RNAs in a sample of saliva without having to resort to a roomful of instruments.
Before this device or any other ambitious application of salivary diagnostics can become a reality, the field as a whole must make significant strides. At present, few salivary biomarkers of disease exist, although we believe that we have promising early evidence that saliva can be used to detect breast cancer, pancreatic cancer and type-II diabetes. But as the field of biomarker diagnosis grows, such molecular signatures may prove to be just the tip of the iceberg—especially if one considers what can also be gleaned from other materials, such as blood, urine, spinal fluid, tears, nipple aspirate or feces. It seems likely that biomarkers for certain diseases will prove to be more abundant in some types of samples than in others. Which works best for what? That question will undoubtedly remain under investigation in coming years. However, my colleagues and I remain optimistic that saliva will continue to reveal secrets about the state of disease or health in both the mouth and the rest of the body. All from lowly spit! We consider it a blessing.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.